Over the past 18 months, significant amounts of data presented at scientific conferences have shed additional light on the mechanisms and clinical significance of antiretroviral drug resistance. These include new reports from studies evaluating the incidence and lingering consequences of transmitted drug-resistant HIV, the significance of the K65R mutation in reverse transcriptase, the persistence of minor HIV variants harboring drug-resistance mutations, the selection of TAM pathways, as well as some heartening data indicating that lamivudine retains some activity against HIV carrying the M184V mutation. To discuss these and other emerging data, Dr. Daniel Kuritzkes graciously accepted PRN’s offer to speak at the May 2004 meeting, the proceedings from which are reviewed here.
| I. Drug-Resistant HIV Transmission | Top of page |
Drug-resistance testing has come to be recognized as an important tool in tracking the growing and troublesome prevalence of transmitted drug-resistant HIV—a problem that appears to be here to stay. To provide PRN members with an update, Dr. Kuritzkes highlighted recent epidemiological data from around the world, looking at different patterns of resistance in both newly infected and chronically infected individuals.
Some of the most intriguing epidemiological data come from the CATCH study, which involved 17 European countries, evaluating the incidence of genotypic resistance in more than 1,600 newly infected HIV-positive individuals (Wensing, 2003). The overall prevalence of HIV strains resistant to at least one antiretroviral agent was 9.6%. “What was interesting about this study was that it was able to divide patients into two groups: those who had been infected for a year or less and those who had been infected for longer than a year, based on the physician’s assessment of infection time,” Dr. Kuritzkes said. According to the data presented at the 2nd International AIDS Society Conference on HIV Pathogenesis and Treatment, held in July 2003 in Paris, the prevalence of drug-resistant HIV among patients infected for a year or less was 10.9%, compared to a prevalence rate of 7.5% among patients infected for more than a year.
Also presented at the 2nd IAS conference were data from the United States, demonstrating similar results. In this study, the prevalence of drug-resistant HIV was assessed among 949 newly infected, antiretroviral-naïve individuals in 10 cities (Bennett, 2003). Using the STAHRS testing algorithm, the investigators determined that 182 patients had been infected within four to six months (primary HIV infection), whereas the remaining 767 patients had been infected for at least six months (chronic HIV infection). Among the patients with primary HIV infection, approximately 11.5% had evidence of resistance to at least one antiretroviral agent. As for patients with chronic HIV infection, approximately 7.5% had evidence of resistance to at least one antiretroviral agent—results similar to the CATCH study. In both studies, resistance to nucleoside reverse transcriptase inhibitors was the most common, followed by resistance to the non-nucleoside reverse transcriptase inhibitors and then the protease inhibitors.
“Not only do the CATCH study and the U.S. study indicate a trend toward increasing prevalence of drug resistance in newly infected individuals,” Dr. Kuritzkes commented, “they also illustrated that drug-resistant HIV persists in plasma for an extended period of time, after transmission has occurred.”
In conflict with the CATCH and U.S.-based studies are data reported at the 11th Conference on Retroviruses and Opportunistic Infections (CROI) suggesting that the prevalence of drug-resistant HIV transmission is actually stabilizing or decreasing. In one study, reporting on new HIV infections at the Academic Medical Center of the University of Amsterdam and the Amsterdam Cohort Studies, the prevalence of transmitted HIV resistant to at least one antiretroviral agent decreased from 20% between the years of 1994 and 1997 to 6% between the years 1998 and 2002 (Bezemer, 2004). And in North Carolina, among the 12 individuals diagnosed with primary HIV infection between January 1998 and June 2000, approximately one-quarter of them had evidence of phenotypic resistance to at least one antiretroviral agent (Hicks, 2004). However, since June 2000, there have been no cases of phenotypic resistance in 18 individuals diagnosed with primary HIV infection.
An explanation for the North Carolina data is not readily available,” Dr. Kuritzkes confessed. “However, it’s important to note that a lot of HIV transmission that has been occurring in recent years is among people of color with limited access to care. And in North Carolina, where the ADAP waiting list is growing longer and longer, it’s possible that the limited care available in the state may be playing a hand in the reduction of transmitted drug-resistant HIV.” As such, differences in the prevalence of resistance, whether between regions or among specific populations, are likely to be important in determining patterns of transmission of resistant HIV.
| III. The Role of Minor Variants | Top of page |
Moving on to resistance issues in heavily pre-treated HIV-positive patients, Dr. Kuritzkes reviewed data stemming from ACTG 398, a clinical trial designed to determine whether the addition of a second protease inhibitor to a regimen containing amprenavir (Agenerase) improved the 24-week response to salvage therapy (Mellors, 2003). The study enrolled 481 heavily pretreated HIV-positive patients: 21% had been on one protease inhibitor in the past, 53% had been on two prior protease inhibitors in the past, and 26% had been on three protease inhibitors in the past. Approximately 44% of the patients had also been on an NNRTI in the past.
All of the patients received efavirenz, adefovir dipivoxil, abacavir, and amprenavir. In addition, subjects were randomized to receive either placebo, nelfinavir (Viracept), saquinavir (Fortovase), or indinavir (Crixivan). There were no statistically significant differences between the three active drug arms. There was, however, a significant difference between the combined active arms and the placebo arm, demonstrating that dual protease inhibitor regimens are superior to single protease inhibitor regimens in heavily pretreated patients.
Not surprisingly, the ACTG 398 investigators reported that NNRTI-naive patients had significantly better responses than NNRTI-experienced patients: 84% of NNRTI-experienced patients, compared to 57% of the NNRTI-naive patients, failed to achieve an undetectable viral load after 24 weeks of follow-up. Of interest, though, is an analysis of the NNRTI-experienced patients. “When we break down the NNRTI-experienced patients into those who did or did not have evidence of NNRTI resistance at baseline, we see that patients who didn’t have evidence of NNRTI resistance seemed to do well during the first 24 weeks of the study,” Dr. Kuritzkes illustrated. After 24 weeks, however, the difference in response rates between these two groups became insignificant and eventually converged. “Why might this be?” Dr. Kuritzkes asked. “There must be something about the prior NNRTI exposure that predisposes this group of patients to fail the NNRTI regimen, compared to patients who have never been exposed to an NNRTI. And whatever that is, it is not being captured using standard resistance assays.”
| Single Genome Sequencing | Top of page |
Commercially available drug-resistance assays involve bulk sequencing, assessing the contribution of all RNA molecules in a plasma sample to generate what is, in essence, an average sequence where minor variants that are present are essentially silenced out by the strong signal from the majority population. An alternative method, Dr. Kuritzkes explained, is to perform single genome sequencing. In single genome sequencing, complementary DNA is derived from plasma RNA—using reverse transcriptase—and is then serially diluted to a single copy. From there, the single copy is amplified and sequenced.
The ACTG 398 study team employed this method to construct a phylogenetic analysis for one patient with NNRTI experience but no evidence of key NNRTI mutations using standard drug-resistance testing at baseline. “This showed that there were 35 sequences from the same patient, the vast majority of which were wild type,” Dr. Kuritzkes explained in his review of the data. “However, two of the 35 sequences carried an NNRTI resistance mutation at codon 108. This is one of the mutations that contributes in a small but significant way to efavirenz resistance. And at the time of treatment failure, the virus that emerged carried the K103N mutation, in addition to the V108I mutation. In other words, the minority variant harboring the V108I mutation had a leg up and started the mutant down the road towards NNRTI resistance and enabled the virus to develop K103N because it was not fully suppressed. This indicates, quite clearly, that these minor variants have a potentially very important role in determining the success of failure for future therapy.”
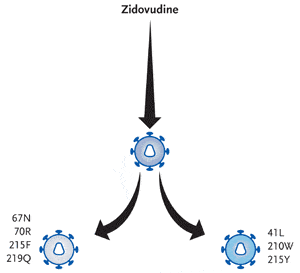
Figure 2. NRTIs exert a blocking effect by plugging a nonextendable nucleoside analogue monophosphate to the 3’ end of the growing proviral HIV-DNA chain. This effectively terminates chain extension and, ultimately, inhibits replication of the virus. However, this process can be reversed by a reverse transcriptase reaction that removes the chain-terminating residue and reinstates an extendable primer. This reverse reaction of DNA polymerization, termed pyrophosphorolysis, enables reverse transcription and DNA synthesis to resume. Pyrophosphorolysis can be enhanced by key mutations, often referred to as thymidine analogue mutations (TAMs), and are the primary mechanism of resistance to zidovudine (Retrovir) and stavudine (Zerit). Several research teams have documented that certain clusters of TAMs develop along different pathways, meaning that some TAMs are more likely to occur together than others. As shown in this figure, the M41L/L210W/T215Y pathway isolate is a common TAM pattern and is associated with high-level resistance to zidovudine and with cross-resistance to other NRTIs, including tenofovir and abacavir. In contrast, the D67N/K70R/K219Q pathway isolate is a less common TAM pattern and is associated with a lower fold resistance than the M41L/L210W/T215Y cluster. Additionally, the T215F mutation rarely appears in the reverse transcriptase of viruses also harboring the L210W mutation or the M41L mutation.
Source: Daniel Kuritzkes, MD
Reviewing some recent work in the area of thymidine analogue mutations, Dr. Kuritzkes explained that certain TAMs tend to occur together but that others are rarely found together in the same virus. For example, the M41L/L210W/T215Y pathway is a common TAM pattern and is associated with high-level resistance to zidovudine and with cross-resistance to other NRTIs, including tenofovir and abacavir (see Figure 2). In contrast, the D67N/K70R/K219Q pathway isolate is a less common TAM pattern and is associated with a lower fold resistance than the M41L/L210W/T215Y cluster. Dr. Kuritzkes also noted that the T215F mutation rarely appears in the reverse transcriptase of viruses also harboring the L210W mutation or the M41L mutation.
Dr. Kuritzkes’ group has conducted fitness and replication studies to better understand these divergent TAM pathways (Hu, 2004). In a nutshell, these studies found that isolates carrying the T215Y mutation replicated more efficiently than isolates harboring the 215F mutation. With incorporation of the L210W mutation into isolates harboring the T215Y mutation, viral fitness was substantially reduced. Conversely, with the incorporation of the K70R mutation into isolates harboring the T215Y mutation, there was a substantial growth advantage in the presence of zidovudine.
“In summary,” offered Dr. Kuritzkes, “we’ve learned that the T215Y pathway is more common and confers higher-level resistance to zidovudine and other NRTIs, whereas viruses carrying T215F have lower replication capacity and are poorly fit.” With respect to the K70R mutation, Dr. Kuritzkes reiterated that it confers a significant advantage to HIV in the presence of zidovudine and that this mutation plays a much larger role in the development of zidovudine resistance than is usually considered.
Perhaps the best known reverse transcriptase mutation is M184V, which is known to cause high-level resistance to lamivudine and emtricitabine. However, there have been studies suggesting that high-level resistance to lamivudine does not necessarily mean that the drug is rendered worthless. An example of this phenomenon can be found in a clinical trial of partial treatment interruptions conducted by Dr. Steven Deeks and his colleagues (Deeks, 2003). This study focused on a cohort of HIV-positive individuals who had a history of excellent treatment adherence, had drug-resistant viremia (greater than 400 copies/mL), and were experiencing a documented treatment-mediated benefit (e.g., a viral load below and CD4+ cell count above pretreatment levels). The patients either stopped their PI(s) or their NRTIs to determine the selective effects of these two drug classes in terms of maintaining the less-fit virus. “To everyone’s surprise,” Dr. Kuritzkes explained, “when the protease inhibitors were stopped in one group of patients, nothing happened. The virus continued to replicate at the same rate without any increase and only a small reemergence of wild-type in the protease gene. This benefit was attributed to the M184V mutation. By contrast, when the nucleoside analogues were stopped in the other group of patients, there was a half to three-quarter log increase in viral load.”
But do these data allow for the conclusion that lamivudine should be maintained in a failing regimen? To explore this further, a team of investigators that included Dr. Kuritzkes discontinued lamivudine in four highly treatment-experienced patients with no viable alternative treatment options and evidence of the M184V mutation while on a regimen consisting of at least three antiretrovirals (Campbell, 2003). Six weeks after stopping lamivudine, HIV-RNA levels increased an average of 0.6 log10 copies/mL above baseline, even though the M184V mutation remained detectable in virus from all patients. “This basically showed us that, even in the presence of the M184V mutation, lamivudine was still contributing to HIV-RNA suppression,” Dr. Kuritzkes explained. Reversion of M184V to wild type occurred in all four patients between six and 14 weeks, accompanied by an additional average increase in viral load of 0.3 log10 copies/mL. Upon resuming lamivudine therapy, the M184V mutation reappeared within eight weeks in all four patients.
However, in another study reviewed by Dr. Kuritzkes, the effects of lamivudine withdrawal in patients with the M184V mutation were less clear. The COLATE trial was an open-label trial involving 131 patients experiencing virologic failure while on a lamivudine-containing regimen (Dragsted, 2004). In switching to another regimen, approximately half of the patients were randomized to continue lamivudine therapy, with the remaining patients randomized to discontinue lamivudine treatment. Forty-eight weeks later, there were no significant differences between the two groups. “However,” Dr. Kuritzkes pointed out, “it’s important to note that this population of patients wasn’t as advanced as our group of patients. In turn, many of the patients responded well to therapy, which ended up swamping the relatively modest effect of residual lamivudine treatment.”
| References | Top of page |
